Protein Secrets Unveiled: New molecular insight of protein–protein interactions
Tokyo, Japan – Proteins are building blocks of our bodies, but they do not work solo. They form partners to facilitate in different biological processes that keep us going. However, analyzing how proteins interact at a molecular level can be challenging. Now, a research team from Japan reveals the secrets behind these “protein partnerships”.
In a study published recently in Protein Science, researchers from Tokyo Medical and Dental University (TMDU) have developed a novel method by combining two techniques in a way that allows identification of the site of interaction on two protein molecules.
A protein consists of a chain of molecules called amino acids, that then forms a three-dimensional structure. An amino acid known as pBpa, which would not naturally occur anywhere else in the protein, can be introduced into a protein of interest using genetic code expansion (GCE). Irradiation with light will then cause two interacting proteins to cross-link and stick together. However, while the site of the pBpa insertion is known, narrowing down the interacting region on the second protein is very difficult.
To solve this, the team introduced a cleavage site, “AllocLys-OH”, at a known location on the second protein using GCE. “Cleaving the crosslinked proteins at this site then provides information as to which side of the AllocLys-OH site the binding interface is located at,” explains first author Kazue Terasawa. “One end of each protein is tagged in a detectable way. If the cleavage experiment separates these tags, then the cleavage site is closer to the tagged end of the protein than the crosslinked interaction site.” By performing a series of these experiments with the AllocLys-OH cleavage site in different positions, the location of the binding interface on the second protein can be narrowed down.
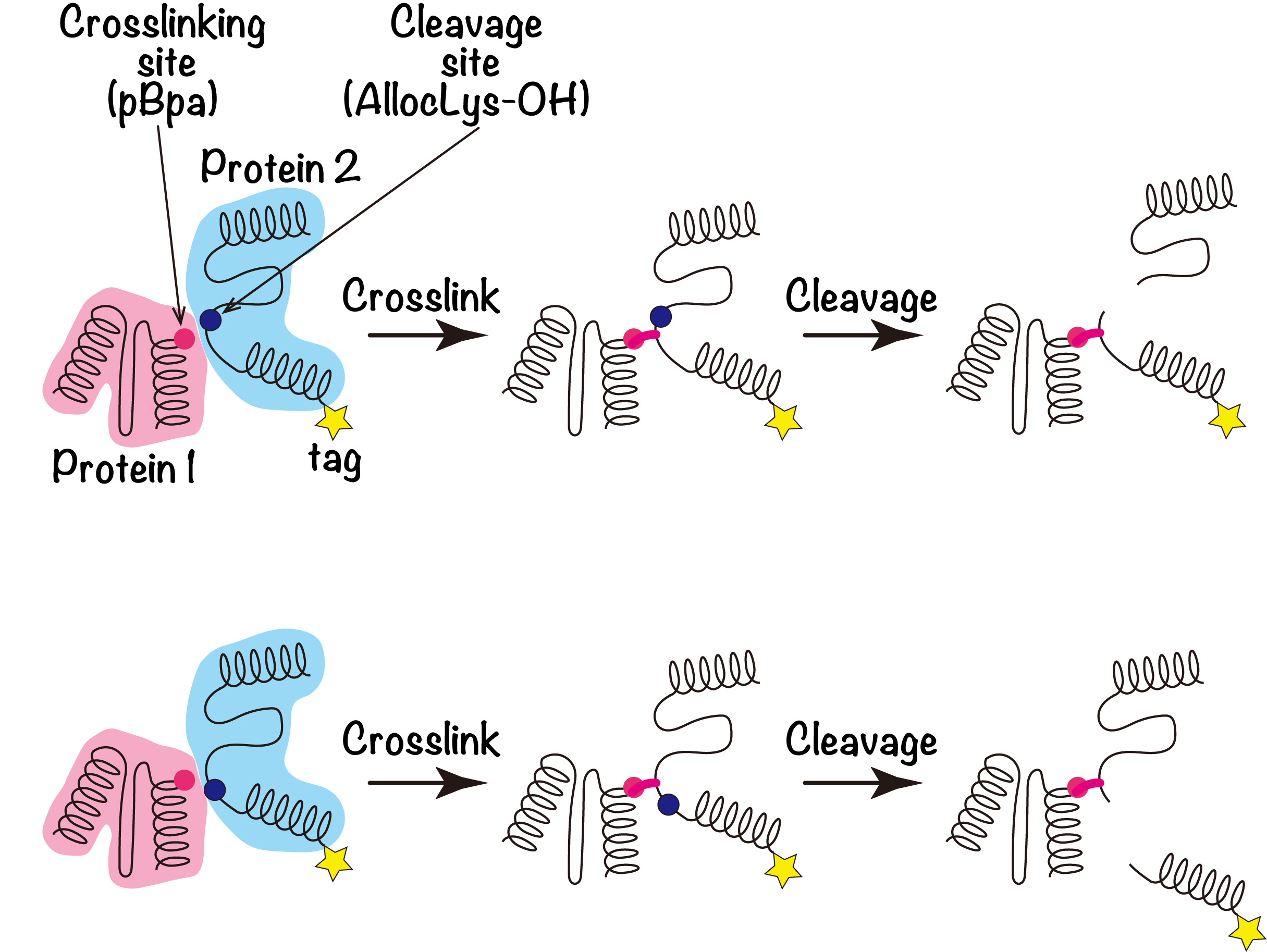
Figure 1. Combination of site-directed photo-crosslinking and AllocLys-OH-mediated site-directed cleavages
The concept of the present method is shown schematically. The crosslinker (pBpa, red circle) or the cleavage site (AllocLys-OH, blue circle) is introduced to Protein 1 or Protein 2, respectively. AllocLys-OH, the α-hydroxy form of AllocLys, forms an alkaline-labile ester bond within the main chain of a protein. After Proteins 1 and 2 are crosslinked, the crosslinked product is cleaved by an alkaline treatment. If the product retains the tag, the crosslinked residue is considered to be closer to the tagged end than the cleavage site in Protein 2 (upper). If the product loses the tag, then the crosslinked residue may be more distal from the tag than the cleavage site (lower).
“After a series of crosslinking and cleavage experiments, we were able to clarify the geometry of the LAMP2A homophilic interaction. Based on the protein’s known three-dimensional structure, the locations of the interaction sites we discovered indicate that LAMP2A must form a trimeric, or higher oligomeric, structure,” explains senior author Miki Hara-Yokoyama.
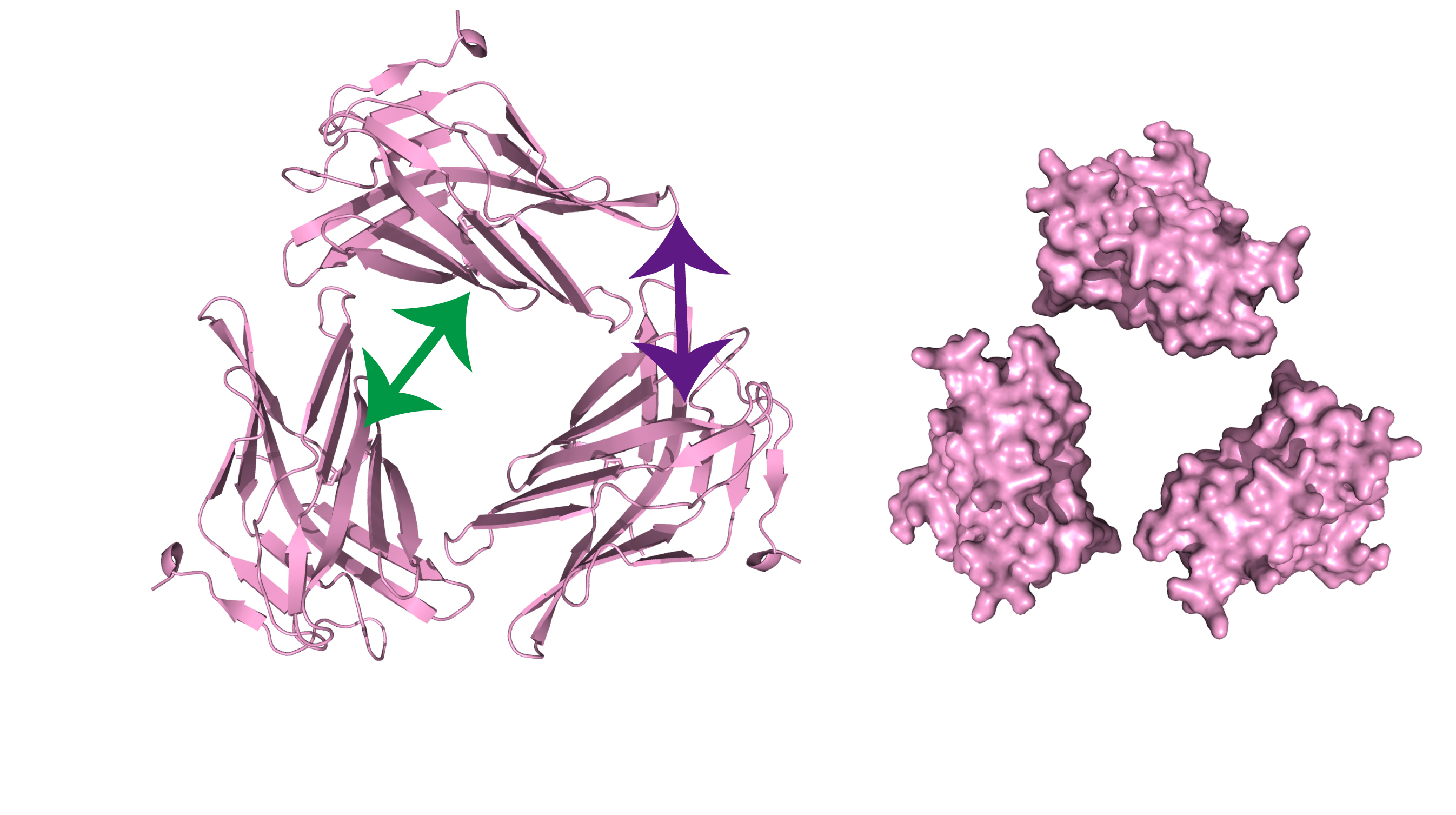
Figure 2. Schematic representation of the trimeric assembly of the C-domain of mouse LAMP2A
The results suggested that at least two adjacent molecules are required to explain the two interactions (shown by green and purple arrows). The presence of the two adjacent molecules is also consistent with a higher oligomeric assembly.
Journal Article
TITLE:Site-specific photo-crosslinking/cleavage for protein–protein interface identification reveals oligomeric assembly of lysosomal-associated membrane protein type 2A in mammalian cells
DOI:https://doi.org/10.1002/pro.4823
Correspondence to
Miki Hara-Yokoyama, Associate Professor
Department of Biochemistry,
Graduate School of Medical and Dental Sciences,
Tokyo Medical and Dental University(TMDU)
E-mail:m.yokoyama.bch(at)tmd.ac.jp
*Please change (at) in e-mail addresses to @ on sending your e-mail to contact personnels.

