Super-Enhancers: The Villain Fueling Certain Cancers
Published: August 5, 2021
Researchers from Tokyo Medical and Dental University (TMDU) identify a regulatory RNA molecule that can help block expression of a powerful cancer-promoting gene
Tokyo, Japan – In a recent study published in CANCER RESEARCH , a team led by researchers at Tokyo Medical and Dental University (TMDU) identified a specific small RNA molecule, called a microRNA (miRNA or miR), that has the potential to be used as an anti-cancer therapeutic. The new research indicates that the molecule, called miR-766-5p, can significantly reduce levels of the oncogene MYC—a specific gene that is expressed at high levels in tumor cells and helps fuel cancer growth and progression.
At its most basic level, cancer is driven by abnormal and uncontrolled gene expression. Numerous different molecular mechanisms contribute to the activation and overexpression of oncogenes in cancer. MiRNAs work as negative regulators of gene expression. This means that they can directly bind and interact with certain gene messages and block them from being translated into a protein. Therefore, any molecular pathway controlled by that specific protein is also affected by this miRNA-mediated regulation. In a previous study, the TMDU group used cell culture experiments to demonstrate that treating cancer cells with miR-766-5p resulted in lower MYC expression and inhibited cancer cell growth rates. Following these intriguing findings, the group aimed to determine the specific mechanism behind these results.
“MYC is a critical oncogene in many tumor types,” says lead author of the study Yasuyuki Gen. “It can promote cancer cell proliferation, can suppress the body’s immune response to fighting the cancer, and can generally be the main driver of tumor progression in many patients.”
The researchers found that miR-766-5p could directly target and reduce expression of two proteins called CBP and BRD4. CBP can induce a molecular change called acetylation that causes DNA to become more “open”, which allows genes present in that area to be more easily expressed. BRD4 can then be recruited to these sites and help promote transcription of these gene messages.
“Areas of DNA with high activity of proteins like CBP and BRD4 are known as super-enhancers,” explains Johji Inazawa, senior author. “Many cancer cells develop super-enhancers near oncogenes, like MYC, that drive increased oncogene expression and therefore promote cancer.”
The team then experimentally treated cells with a synthetic version of miR-766-5p, finding that the resulting suppression of CBP and BRD4 caused decreased MYC levels in cancer cells, but not in normal cells. Additionally, tumors that were engrafted in lab mice showed significantly suppressed growth when treated with miR-766-5p compared with a control miRNA.
“Our findings suggest that miR-766-5p-mediated control of CBP and BRD4 blocks formation of the super-enhancers that contribute to MYC overexpression in cancer cells,” explains Gen.
In recent years, efforts have been made to develop specific miRNAs into targeted therapies for various cancers. This study provides considerable evidence that miR-766-5p could be used to fight MYC-driven cancers by targeting super-enhancers.
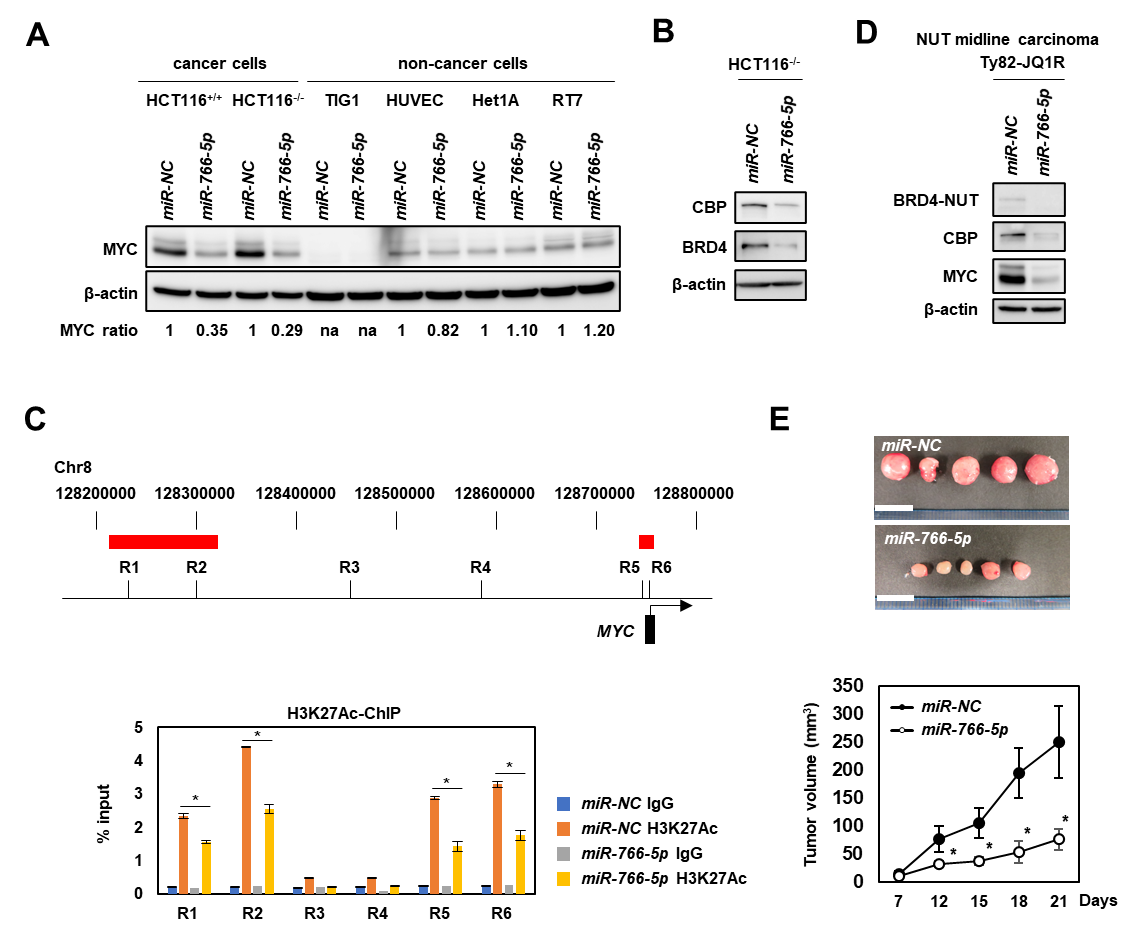
Figure 1. Tumor suppressive functions of miR-766-5p
(A) Western blot analysis of MYC in indicated cell lines after transfection with 10 nmol/L of miR-NC or miR-766-5p. The intensity of MYC bands was quantified by densitometry and shown as the fold change after normalization with β-actin. na, not applicable. (B) Western blot analysis of CBP and BRD4 in HCT116-/- cells 48 hours after transfection with 10 nmol/L of miR-NC, or miR-766-5p. (C) H3K27Ac Chromatin immunoprecipitation (ChIP)-qPCR at the indicated loci in HCT116-/- cells transfected with miR-NC or miR-766-5p. Red bars indicate super-enhancer regions of MYC. Relative ChIP enrichment values in the indicated regions are expressed as percentages relative to input DNA. Bar, SD for triplicate experiments. * P < 0.05. (D) Western blot analysis of BRD4-NUT, CBP, and MYC expression in Ty82-JQ1R cells transfected with 10 nmol/L of miR-NC or miR-766-5p. (E) Local administration of miR-766-5p suppressed in vivo tumor growth. (upper) Resected tumors at 21 days after the injection of Ty82-JQ1R cells. Scale bar, 10 mm. (lower) Tumor growth curves of xenograft mouse models treated with miR-NC or miR-766-5p (n = 5, each).
(A) Western blot analysis of MYC in indicated cell lines after transfection with 10 nmol/L of miR-NC or miR-766-5p. The intensity of MYC bands was quantified by densitometry and shown as the fold change after normalization with β-actin. na, not applicable. (B) Western blot analysis of CBP and BRD4 in HCT116-/- cells 48 hours after transfection with 10 nmol/L of miR-NC, or miR-766-5p. (C) H3K27Ac Chromatin immunoprecipitation (ChIP)-qPCR at the indicated loci in HCT116-/- cells transfected with miR-NC or miR-766-5p. Red bars indicate super-enhancer regions of MYC. Relative ChIP enrichment values in the indicated regions are expressed as percentages relative to input DNA. Bar, SD for triplicate experiments. * P < 0.05. (D) Western blot analysis of BRD4-NUT, CBP, and MYC expression in Ty82-JQ1R cells transfected with 10 nmol/L of miR-NC or miR-766-5p. (E) Local administration of miR-766-5p suppressed in vivo tumor growth. (upper) Resected tumors at 21 days after the injection of Ty82-JQ1R cells. Scale bar, 10 mm. (lower) Tumor growth curves of xenograft mouse models treated with miR-NC or miR-766-5p (n = 5, each).
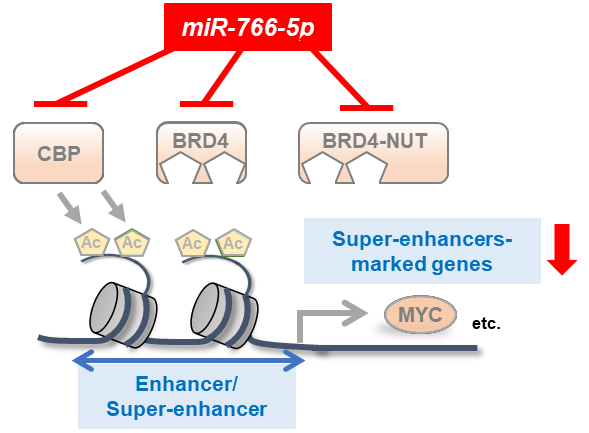
Figure 2. Diagram summarizing the mechanism by which miR-766-5p reduces the activity of super-enhancers in cancer cells.
miR-766-5p downregulated the levels of H3K27Ac at super-enhancer regions by targeting acetyl transferase CBP. H3K27Ac is required for enhancer activation and BRD4 binds to active enhancers to drive transcription. As miR-766-5p concurrently targets CBP and BRD4 or BRD4-NUT, miR-766-5p may efficiently suppress the expression of SE-marked genes such as MYC in cancer cells.
miR-766-5p downregulated the levels of H3K27Ac at super-enhancer regions by targeting acetyl transferase CBP. H3K27Ac is required for enhancer activation and BRD4 binds to active enhancers to drive transcription. As miR-766-5p concurrently targets CBP and BRD4 or BRD4-NUT, miR-766-5p may efficiently suppress the expression of SE-marked genes such as MYC in cancer cells.
###
The article, “miR-766-5p targets super-enhancers by downregulating CBP and BRD4,” was published in CANCER RESEARCH at DOI: 10.1158/0008-5472.CAN-21-0649Summary
Researchers from Tokyo Medical and Dental University (TMDU) identified a small RNA molecule called miR-766-5p that reduces expression of MYC, a critical cancer-promoting gene. This microRNA reduces levels of proteins CBP and BRD4, which are both involved in super-enhancer (SE) formation. SEs form in areas of DNA that can fuel MYC expression and tumor progression. This study provides strong evidence for developing miR-766-5p as a novel therapeutic to treat MYC-driven cancers.
Journal Article
JOURNAL:Cancer Research
TITLE:miR-766-5p targets super-enhancers by downregulating CBP and BRD4
DOI:https://doi.org/10.1158/0008-5472.CAN-21-0649
TITLE:miR-766-5p targets super-enhancers by downregulating CBP and BRD4
DOI:https://doi.org/10.1158/0008-5472.CAN-21-0649
Correspondence to
Yasuyuki Gen,Assistant Professor
Department of Molecular Cytogenetics,
Medical Research Institute,
Tokyo Medical and Dental University(TMDU)
E-mail:johinaz.cgen (at) mri.tmd.ac.jp
E-mail:ygen.cgen (at) mri.tmd.ac.jp
*Please change (at) in e-mail addresses to @ on sending your e-mail to contact personnels.

