”Role for Autophagic Cellular Degradation Process in Maintaining Genomic Stability”

Shigeomi SHIMIZU, MD, PhD, Professor
Department of Pathological Cell Biology, Medical Research Institute, TMDU (Right)
Shinya HONDA, PhD, Assistant Professor
Department of Pathological Cell Biology, Medical Research Institute, TMDU (Left)
Tokyo, Japan – Centrosomes play an essential role in cell division by organizing the protein framework on which chromosomes assemble and then separate prior to division into daughter cells. Centrosomes are made up of a pair of centrioles, which are themselves composed of different proteins such as centrosomal protein 63 (Cep63) and polo-like kinase 4. These proteins are thought to regulate the number of centrioles, and thus centrosomes as well, using the ubiquitin–proteasome breakdown pathway that first adds a molecular tag to the protein to be degraded. However, researchers centered at Tokyo Medical and Dental University observed that mouse cells lacking a protein involved in autophagy (a lysosome-based degradation process involving hydrolytic enzymes) contained multiple centrosomes. Their work showed that autophagy is another method of regulating centrosome number. The study was reported in Nature Communications.
Selective autophagy has been shown to break down specific cargos, including proteins and organelles, by delivering them to the lysozyme for digestion. The research team found that cells deficient in autophagy carry extra centrosomes, which was confirmed by the use of autophagy inhibitors in normal cells.
Multiple dots of Cep63 were also visible in cells not undergoing autophagy. Silencing of Cep63 reduced the population of cells with multiple centrosomes while its upregulation increased the centrosome number, thus confirming a role for Cep63 in centrosome number regulation. “Protein staining showed that the Cep63 dots co-localize and directly associate with the p62 protein, which plays a role as a cargo receptor in selective autophagy”, first author Yuichiro Watanabe says. “This revealed that p62 is involved in the autophagic degradation of Cep63.” In normal cells, most Cep63 dots undergo autophagy to regulate the centrosome number. However, in the absence of autophagy, the Cep63 dots persist and interact with p62 or lead to the generation of additional centrosomes.
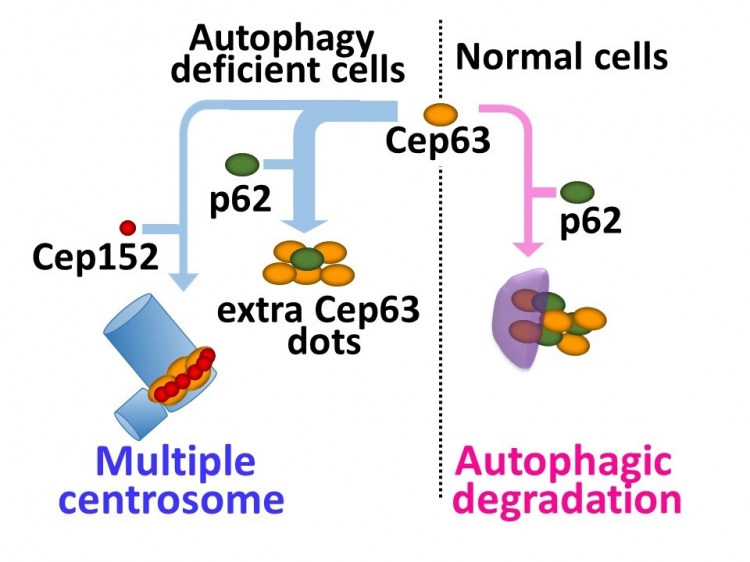
Fig Autophagy controls centrosome number by degrading Cep63
The ubiquitin-proteasome system is thought to be the primary regulator of centrosome number. Here, Watanabe et al. show that selective autophagy also plays a role in regulating centrosome number via p62-dependent recruitment of centrosomal protein 63 to autophagosomes.
The article “Autophagy controls centrosome number by degrading Cep63” was published in Nature Communications at doi:10.1038/ncomms13508.
Summary text:
Correspondence to:
Department of Pathological Cell Biology,
Medical Research Institute,
Tokyo Medical and Dental University (TMDU)
E-mail: shimizu.pcb(at)mri.tmd.ac.jp

