Integrating Data to Find a Cure
Published: August 17, 2021
Researchers from Tokyo Medical and Dental University (TMDU) use a comprehensive analytical approach to uncover molecular characteristics of esophageal cancer in Japanese patients
Tokyo, Japan – Abnormal gene expression is ultimately what fuels cancer. Certain expression profiles or signatures can help physicians diagnose an individual with a specific type of cancer. However, gene expression analysis can also be used to identify new targets for anti-cancer therapeutics.
In a recent study published in Cancer Science, a team led by researchers at Tokyo Medical and Dental University (TMDU) performed several analyses on esophageal squamous cell carcinoma (ESCC) tumors from Japanese patients. They identified specific variations and alterations in genes commonly found in ESCC patients. Identifying the genes that promote the development and progression of ESCC is one step towards improving our understanding of how to treat this disease.
ESCC is the predominant form of esophageal cancer in Asia and remains a deadly disease in need of innovative treatment methods. An effective targeted therapy for ESCC has yet to be developed. Previous studies have identified specific genetic variants associated with ESCC. Scientists have also examined DNA methylation patterns, which are a form of epigenetic modification. When certain regions of a DNA molecule become methylated, gene expression levels can change. Yet these patterns have mostly been used as biomarkers to identify ESCC subtypes. Because of this, the TMDU group aimed to integrate epigenetic data with DNA sequencing and gene expression trends to better understand ESCC.
“Various molecular events simultaneously contribute to cancer development,” says lead author of the study Akira Takemoto. “Our new integrative approach can help reveal the gene variants, pathways, and altered functions that are common in Japanese ESCC patients.”
The analysis suggested that many ESCC tumors displayed variants of certain cancer-related genes, such as members of the NOTCH family and the tumor suppressor gene TP53. A variant of TP53 was detected in 84% of the tumor samples. Additionally, some of these genes showed copy number amplification.
“We also examined cancer-related genes that display allelic expression imbalances (AEIs), meaning one allele is predominantly expressed,” states Johji Inazawa, senior author. The team discovered that many ESCC patients had AEI in gene members of the FAT family. Some cases had highly expressed alleles with variants, which could support tumor growth. “Humans have two alleles for each gene, one inherited from their mother and another from their father. AEIs can affect the expression balance between these alleles.”
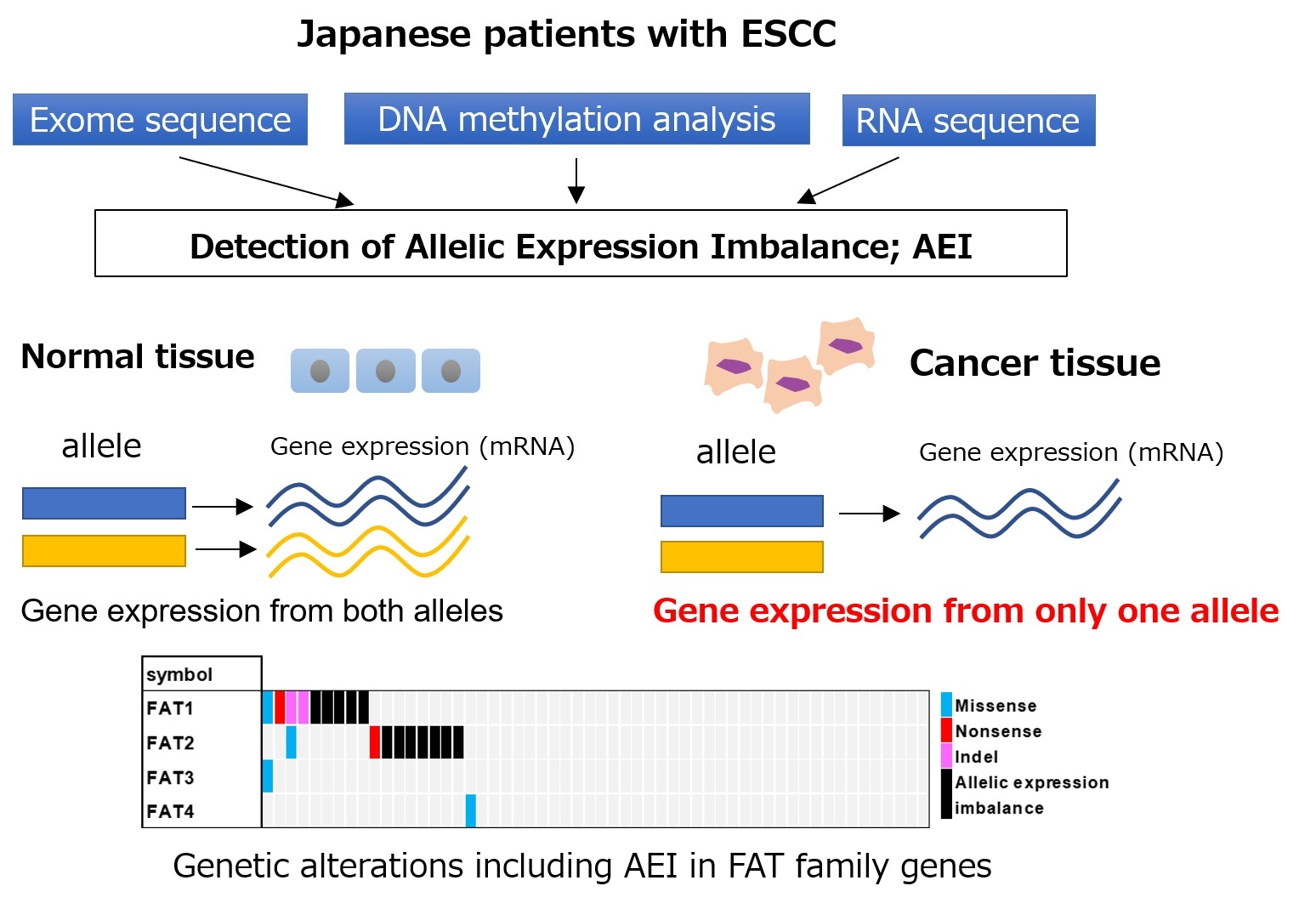
Figure. AEI occurring in ESCC
In previous studies, several cancer related genes were found to be involved in AEIs, which expressed predominantly only one allele of two.Our integrated analyses revealed genome-wide profiles of AEIs in ESCCs.
In addition, approximately 30% of ESCC cases with nonsynonymous SNVs or AEI in FAT1 and FAT2 genes, which are well-known to be mutated in cancer, tended to be detected in a mutually exclusive manner.
“Overall, our findings reveal the molecular landscape of ESCC in Japanese patients, including data on mutations, gene expression levels, signaling pathways, and DNA methylation,” explains Takemoto.
The intriguing results generated by this team provide a more comprehensive view of ESCC tumors. This work will be key for discovering new targets and developing effective therapies for this deadly disease.
###
The article, “Integrative genome-wide analyses reveal the transcriptional aberrations in Japanese esophageal squamous cell carcinoma,” was published in Cancer Science at DOI: 10.1111/cas.15063.Summary
Researchers from Tokyo Medical and Dental University (TMDU) performed an integrative analysis of gene sequences, DNA modification patterns, and gene expression levels in esophageal squamous cell carcinoma (ESCC) tumors from Japanese patients to uncover potential molecular signatures of this disease. They identified gene variants common in ESCC and determined specific pathway and gene functions frequently altered in these tumors. These results may provide potential new targets for other therapeutic interventions for ESCC.
Journal Article
JOURNAL:Cancer Science
TITLE:Integrative genome-wide analyses reveal the transcriptional aberrations in Japanese esophageal squamous cell carcinoma
DOI:https://doi.org/10.1111/cas.15063
TITLE:Integrative genome-wide analyses reveal the transcriptional aberrations in Japanese esophageal squamous cell carcinoma
DOI:https://doi.org/10.1111/cas.15063
Correspondence to
Department of Molecular Cytogenetics,
Medical Research Institute,
Tokyo Medical and Dental University(TMDU)
E-mail:johinaz.cgen (at) mri.tmd.ac.jp
Kousuke Tanimoto, Assistant Professor
Genome Laboratory,
Medical Research Institute,
Tokyo Medical and Dental University(TMDU)
E-mail:ktani.nri (at) mri.tmd.ac.jp
*Please change (at) in e-mail addresses to @ on sending your e-mail to contact personnels.

