"Differences in Microbial Interactions Underlie Related Oral Diseases"
Tokyo – Microorganisms including bacteria, fungi, and viruses coexist on or within the human body, often as multifaceted polymicrobial biofilm communities. Most of infectious diseases are caused by a mixture of microbes, and an understanding of the ability of microorganisms to cause these polymicrobial diseases is crucial for treatment and prevention. However, this has been challenged by similarities between symptoms of different polymicrobial diseases yet differences in their response to treatment.
Now, Japanese researchers led by Tokyo Medical and Dental University (TMDU) have examined the interactions and biochemical functions of microbial communities to help discriminate between two similar oral diseases. The study was reported in Scientific Reports.
Periodontitis is an inflammatory polymicrobial disease of the gums and associated with systemic disease such as cardiovascular disease and diabetes. Peri-implantitis shares several symptoms with periodontitis but responds less well to treatment. It is caused by a polymicrobial infection of tissues surrounding dental implants and often leads to loss of the implant. Previous studies investigated the microbial species found in both diseases by sequencing DNA fragments, including those encoding a gene (16S rDNA) that evolves very slowly so can be used to study evolutionary relationships. However, results were conflicting, perhaps reflecting the fact that samples were obtained from different individuals or may have included dead or inactive microbes.
To overcome this, TMDU researchers isolated microbial samples from 12 adults who had both periodontitis and peri-implantitis to eliminate potential differences between individuals. They then classified the microbes by assessing the active transcription of the 16S rDNA gene to exclude dead species. This revealed presence of microbial species between periodontitis and peri-implantitis samples.
Despite these differences, the team observed several functional similarities in the roles of the microbial communities following an analysis of their genetic activity (Figure1). Using several protein databases to identify gene functions, the two sample types were found to have shared use of metabolic pathways and virulence factors (molecules that enable pathogenicity such as replication or evading the host immune system).
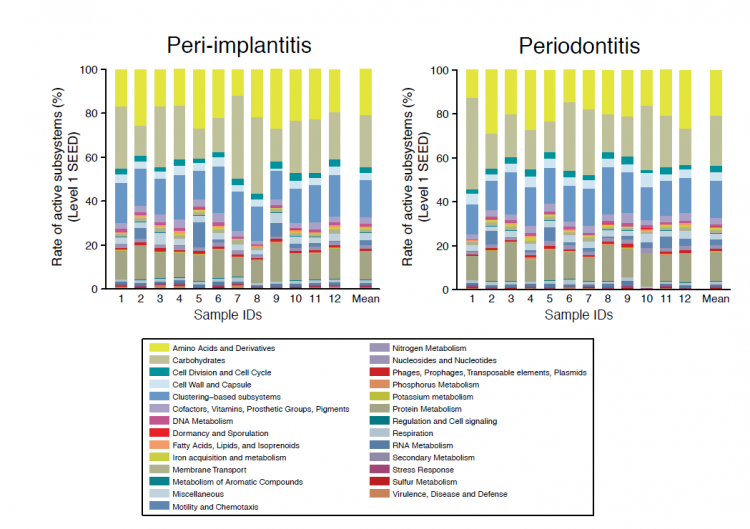
Fig1: Comparison of functional profiles of peri-implantitis and periodontitis.
The composition of functional genes appear similar among the samples for each disease. The subsystems of “carbohydrates” and “protein metabolism” are predominant in both peri-implantitis and periodontitis sites.
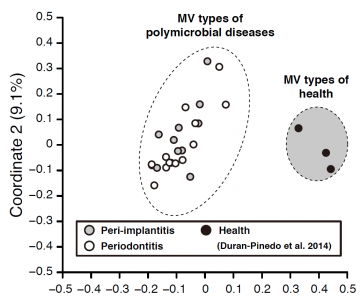
Fig2: PCoA plots of microbial virulence types (MV types) between polymicrobial disease and health.
The MV types of the polymicrobial diseases (peri-implantitis and periodontitis) were apparently dissimilar with those of healthy sites. MV types made only by assignments with virulence factors can serve as clear indicators to distinguish between samples from diseased and healthy states. While, the MV types of the peri-implantitis and periodontitis sites were likely similar, indicating that both diseases might be based on similar virulence functions.
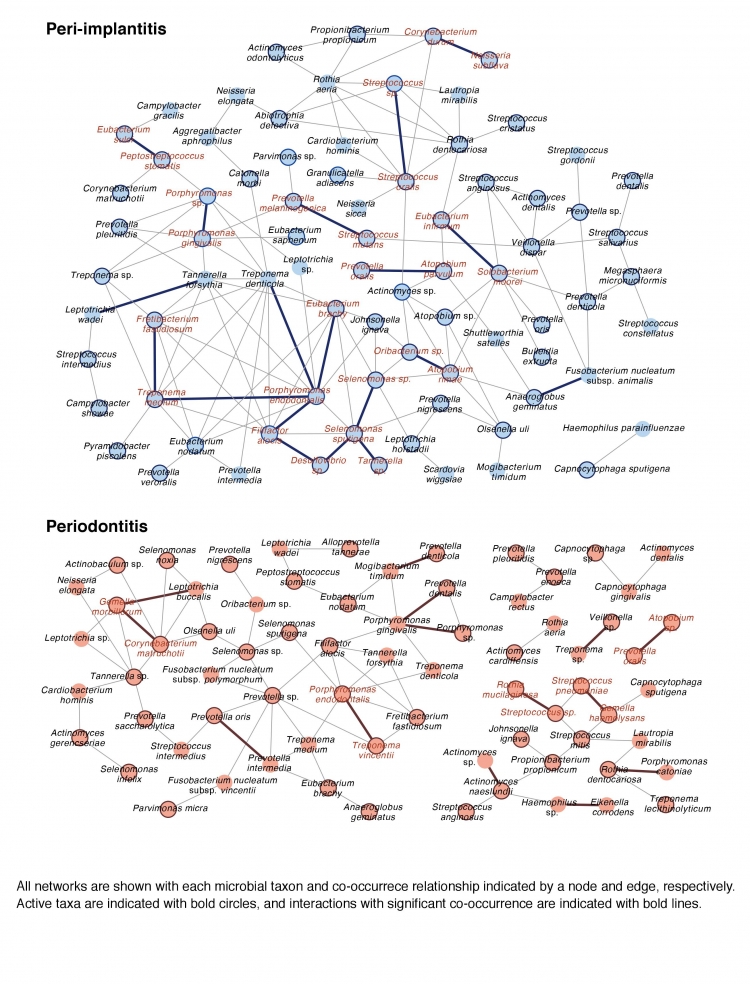
Fig3:Co-occurrence networks of microbial taxon in peri-implantitis and periodontitis.
Some of active species with high transcriptional activity are connected with significant co-occurrence and these taxa are detected in more than two-thirds of patients, as “interacting core taxa”. Interacting core taxa are indicated in red text for peri-implantitis and periodontitis. We propose that dissimilarities of interacting core taxa in co-occurrence networks and network structure can cause differences in the etiologies of both disease.
Summary Text:
Correspondence to:
Department of Periodontology,
Graduate School of Medical and Dental Sciences,
Tokyo Medical and Dental University(TMDU)
E-mail: takeuchi.peri(at)tmd.ac.jp

