Genomics in cancer research leads to improved prognoses and new therapies

Shumpei Ishikawa
Professor of Genomic Pathology
Profile
Dr. Ishikawa graduated from the Faculty of Medicine at the University of Tokyo in 2000, and received his PhD degree in Pathology. He performed postdoctoral research in the Genome Science Division of the Research Center for Advanced Science and Technology at the University of Tokyo. He became an Associate Professor in the Department of Pathology there, and joined TMDU as a Professor of Genomic Pathology in 2013.
PDF Download
- Genomics in cancer research leads to improved prognoses and new therapies (PDF: 642KB)
A: We are working on any intractable diseases that have no effective therapeutics. Diffuse-type gastric cancer is one such disease, with a strong impact on public health in Japan. It is usually said that cancer is a disease for elderly people, but diffuse-type gastric carcinoma is common at relatively younger ages, so its effect on economic activity is significant. Genome sequencing is a powerful approach for profiling somatic mutations, but in addition to simple cancer cell abnormality, we also focus on multicellular complex systems like cancer-stromal interaction and cancer immunology. We are taking a data-science approach, gathering a large amount of genomic data, which can lead to the understanding of disease systems and identification of therapeutic targets and biomarkers.
A: In cancer genome analysis, two type of somatic mutations are observed: mutations of driver genes, which are important for cancer development and progression, and passenger gene mutations, which are stochastically incorporated into the cancer genome and are not important for the cancer cell. To discover a small number of true driver gene mutations among a large number of background candidate mutations, we collect genome sequencing data from many cases and focus on the mutation density and distribution within genes. In particular, the mutation positions in relation to the protein structure and the amino acid substitution pattern are important. Knowledge and experience of bioinformatics and structural biology are needed. It is also necessary to validate the biological significance of our data-based findings by combining them with laboratory experiments.
A: Diffuse-type gastric cancer has a poor prognosis because currently there are no therapeutics available. Our research shows that mutant RHOA is a driver mutation that is indispensable for the disease phenotype of diffuse-type gastric cancer. It is possible that a molecular-targeted drug can be successfully developed for mutant RHOA. With current drug design technology, only limited protein families, such as the kinase family, can be targeted, and these are called “druggable proteins”. Mutant RHOA is not a typical “druggable” protein, so further knowledge of its exact biochemical mechanism is needed in order to discover a “druggable” point. New drug development technologies, such as simulation of protein-drug interaction and DDS (Drug Delivery Systems) for nucleic acid drugs will be also necessary.
A: In this Center, we collect and deposit clinical materials from TMDU hospital patients with their informed consent. I am working on the development of a processing protocol for disease tissue used in genomics research. Unlike the case of peripheral blood, the preservation and nucleic acid extraction protocols vary for disease tissue, and could differ depending on disease type and locus. The differences in sample-by-sample and inter-facility bioresource quality has proven to be a significant problem for genomics research, and a standardized process is necessary to address this issue.
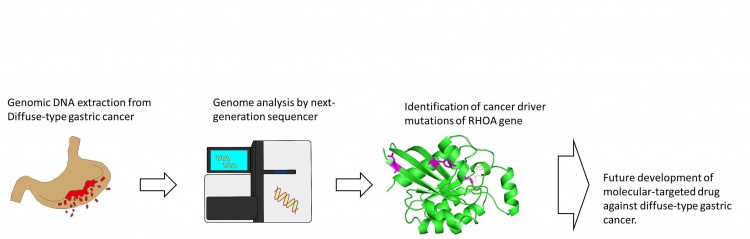
Genome sequencing and therapeutic target discovery for diffuse-type gastric cancer.
A: I am eager to collaborate in research on cancer genomics and immunogenomics using Bioresource Research Center materials. In particular, oral cancer is strong area of TMDU and excellent research resources exists. As to international collaboration, we are now conducting research with Indian groups about functional genomics screening for Indian anti-tumor natural products. In addition to disease research and clinical diagnosis, genomics could be applied to microbe metagenomics, functional genomics screening, and synthetic biology, which are among our strong interests. Also, genome science has an important role in social and medical infrastructure, so I would like to contribute to Japanese society by contributing to the creation of national policy in this area.
Journal Information
Nat Genet. ,doi: 10.1038/ng.2984.

