Dynamic molecular network analysis of iPSC-Purkinje cells differentiation delineates roles of ISG15 in SCA1 at the earliest stage Abstract Journal
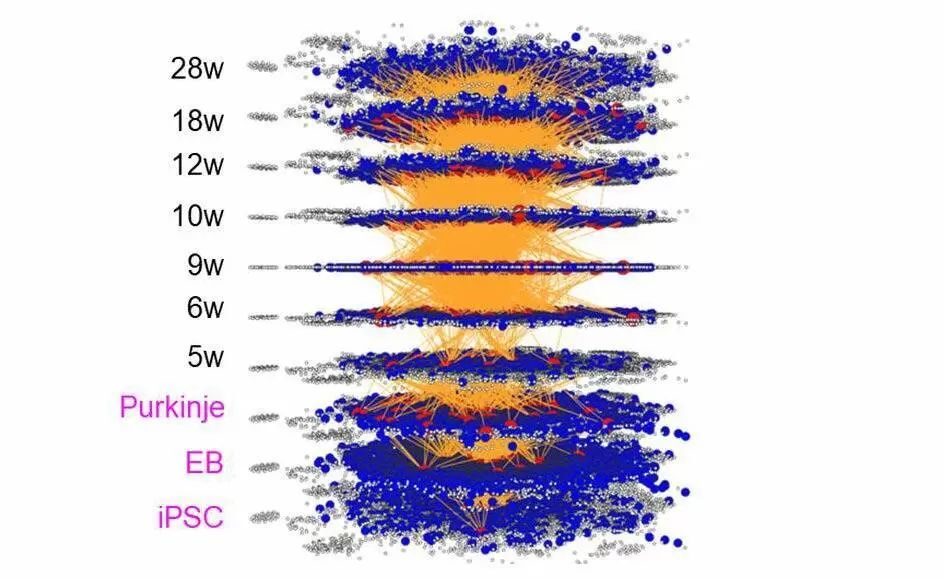
A simulation result of iMAD, a new method of dynamic molecular network analysis. This method can visualize "cause-result relationships" of chronological changes of molecular networks.
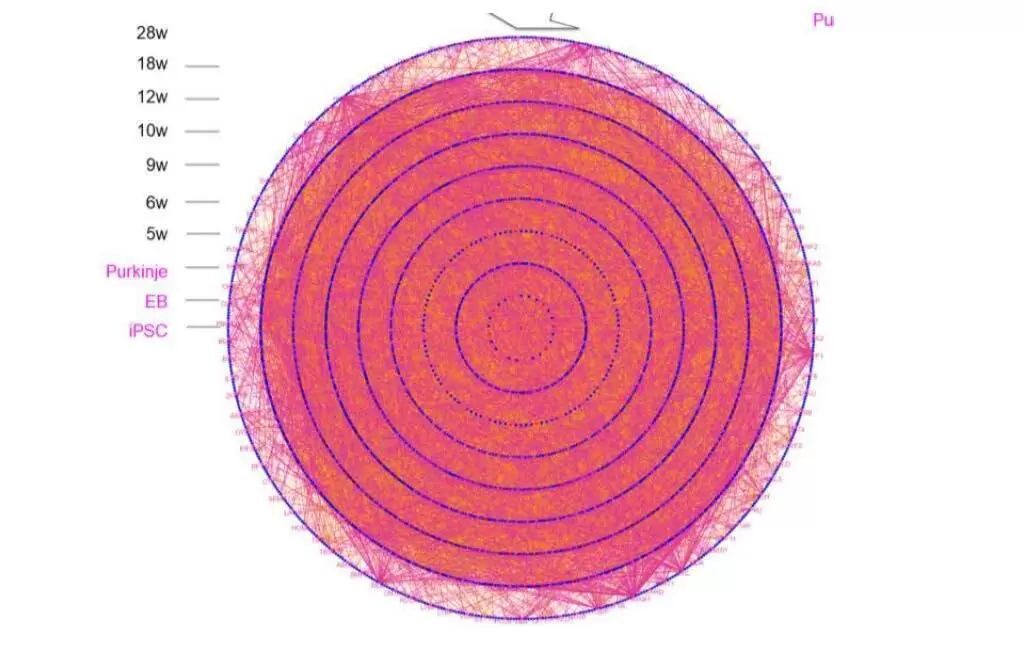
Purple lines show a cytokine-relevant molecular network connected from iPSCs stage to 18 months of age of mice.
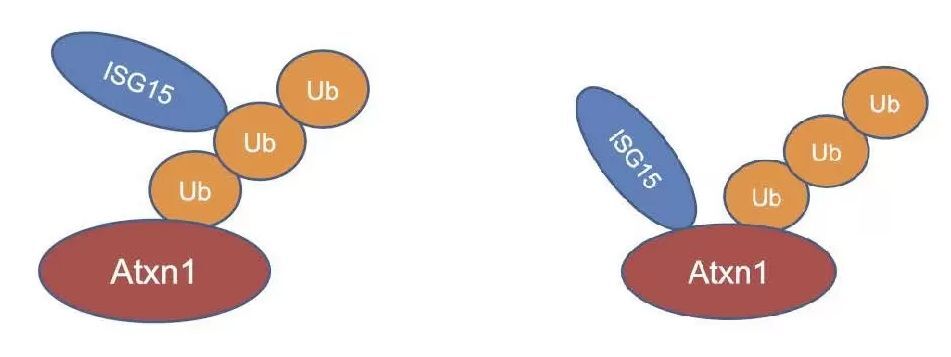
ISG15 inhibites proteolysis by binding to poly-ubituitin on Atxn1 protein directory (left), or ubiquitinated Atxn1 proteins(right).
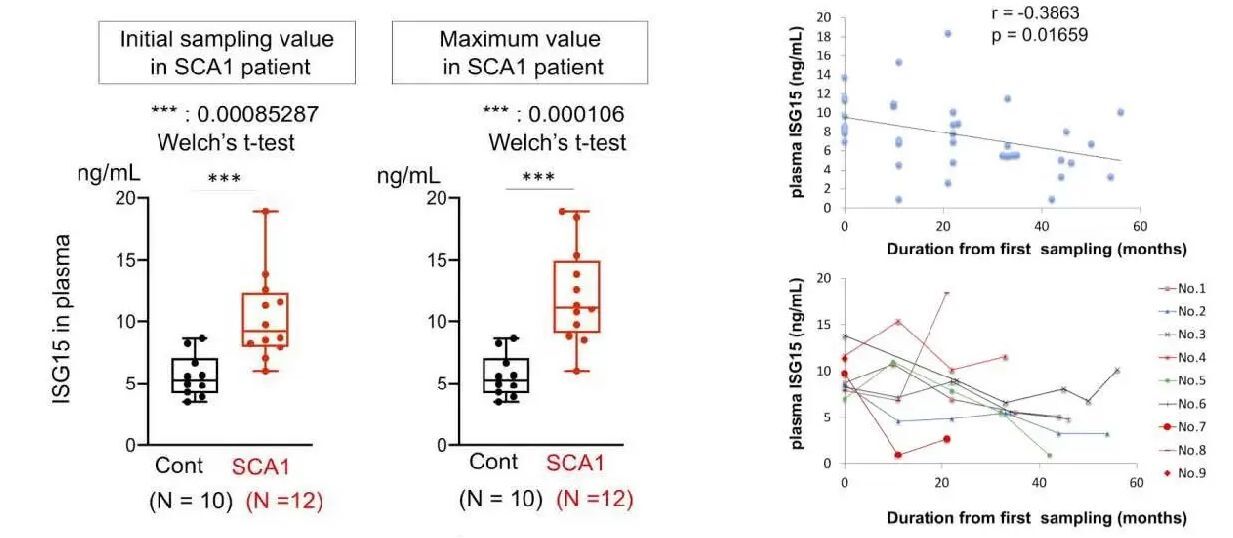
ISG15 levels in plasma of non-neurological disease controls or SCA1 patients.
Publications
Dynamic molecular network analysis of iPSC-Purkinje cells differentiation delineates roles of ISG15 in SCA1 at the earliest stage.
Commun. Biol. 9 April 2024, 7 (1), 413. doi: 10.1038/s42003-024-06066-z
Related contents
- TMDU press release - "Dynamic molecular network analysis of iPSC-Purkinje cells differentiation delineates roles of ISG15 in SCA1 at the earliest stage"
- Springer Nature Research Communities - "How we can simulate temporal changes of molecular networks based on the cause-result relationship?"

