Research Interests
Introduction
Alternative splicing of precursor mRNA (pre-mRNA) is an important mechanism for producing proteome diversity in multicellular organisms.
Recent high-throughput sequencing analysis of human tissue transcriptomes revealed that more than 90% of human genes undergo alternative splicing and that most of these alternative splicing events vary between tissues.
Alternative pre-mRNA processing can be classified according to seven basic elements involved in such events: cassette exons, which are included in certain mRNA isoforms and skipped in others; mutually exclusive exons; alternative 5′ splice sites; alternative 3′ splice sites; intron retention; alternative promoters; and alternative poly(A) sites.
The mechanisms involved in regulating alternative splicing in living cells have, in the past, been studied using splicing-reporter minigenes consisting of multiple exons and introns; alternative mRNA isoforms derived from reporter minigenes were analyzed by quantifying reverse transcription PCR (RT-PCR) products after isolating total RNA from transfected cells. However, the laborious nature of these procedures prevented high-throughput analysis of alternative splicing regulation in living cells or organisms.
Recent high-throughput sequencing analysis of human tissue transcriptomes revealed that more than 90% of human genes undergo alternative splicing and that most of these alternative splicing events vary between tissues.
Alternative pre-mRNA processing can be classified according to seven basic elements involved in such events: cassette exons, which are included in certain mRNA isoforms and skipped in others; mutually exclusive exons; alternative 5′ splice sites; alternative 3′ splice sites; intron retention; alternative promoters; and alternative poly(A) sites.
The mechanisms involved in regulating alternative splicing in living cells have, in the past, been studied using splicing-reporter minigenes consisting of multiple exons and introns; alternative mRNA isoforms derived from reporter minigenes were analyzed by quantifying reverse transcription PCR (RT-PCR) products after isolating total RNA from transfected cells. However, the laborious nature of these procedures prevented high-throughput analysis of alternative splicing regulation in living cells or organisms.
A Transgenic Reporter Worm System Offers a Path to Alternative Splicing Codes in vivo
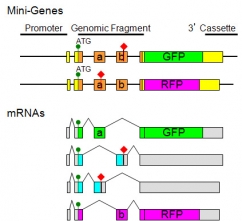
We have recently utilized C. elegans as a model organism to study alternative splicing regulation in vivo (Nat Meth, 2006; Nat Protoc, 2010). By constructing multi-chromatic alternative splicing reporter mini-genes for C. elegans, we successfully visualized cell-type-specific and developmentally regulated alternative splicing events in vivo (Mol Cell Biol, 2007; Genes Dev, 2008).
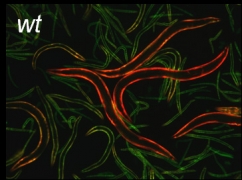
Developmental regulation of the let-2 alternative splicing reporter.
As C. elegans is transparent, it is easy to observe expression patterns of multiple fluorescent proteins in living worms at a single cell resolution. Furthermore, a variety of genetic tools for C. elegans research, such as transgenic expression of exogenous proteins, mutant screening and gene mapping, and RNAi-mediated gene knock-down, facilitated further analyses of trans-acting factors and cis-elements, and identification of partially spliced RNA species.
Another advantage of C. elegans in studying splicing regulation is that its introns are on average very short and therefore it is easy to construct reporter mini-genes that include all the required elements.
The studies on splicing regulation in C. elegans revealed that regulation mechanisms of alternative splicing is evolutionarily conserved between nematodes and mammals, and therefore further studies in C. elegans will help determine cellular codes for alternative splicing in higher organisms. Our reporter system will further elucidate expression profiles and regulation mechanisms of alternative splicing in vivo.
Another advantage of C. elegans in studying splicing regulation is that its introns are on average very short and therefore it is easy to construct reporter mini-genes that include all the required elements.
The studies on splicing regulation in C. elegans revealed that regulation mechanisms of alternative splicing is evolutionarily conserved between nematodes and mammals, and therefore further studies in C. elegans will help determine cellular codes for alternative splicing in higher organisms. Our reporter system will further elucidate expression profiles and regulation mechanisms of alternative splicing in vivo.
Former Professor, Dr. Masatoshi HAGIWARA, and his group members moved to Kyoto University in summer, 2010.

