|
 |

2. Regulating mechanism of alternative splicing and its physiological function during the development of mouse brain
Alternative splicing generates a large number of mRNA and protein isoforms from a limited number of gene subsets. In the brain, many important molecules are regulated their structures and functions with this system, like signaling receptors, cell adhesion molecules, transcription factors, and so on. This mechanism contributes to the functional complexity and high performance of the mammalian brain through making divergent protein subsets from small number of genes. Also many neuronal diseases are caused by the mis-regulation of alternative splicing, i.e.; neurodegenerative disease, mental disorder, neuron-muscular dystrophy, or brain tumors. However, the mechanism of gene-, cell-, organ-, developmental stage-, and brain structure-specific regulation of alternative splicing is still mostly unknown.
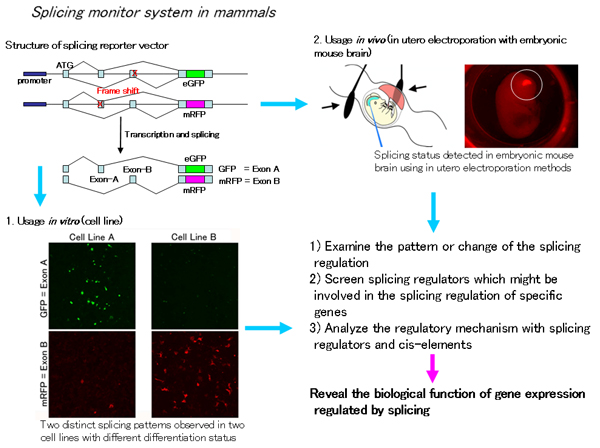
We are developing a monitoring system of alternative splicing during the development of the brain using a fluorescent-based reporter vector system, and revealing their dynamism in single cell resolution. With this system, we screen and select molecules that are essential for regulating alternative splicing, and examine their biological function during development. This project will reveal the regulating mechanism of alternative splicing and its important roles from in vitro to in vivo
3. mRNA splicing regulation and virus infection.
Although the viral genome is often quite small, it encodes a broad series of proteins. The virus takes advantage of the host-RNA-processing machinery to provide the alternative splicing capability necessary for the expression of this proteomic diversity. Serine-arginine-rich (SR) proteins and the kinases that activate them are central to this alternative splicing machinery. We originally developed SR protein phosphorylation inhibitor 340 (SRPIN340), which preferentially inhibits SRPK1 and SRPK2. SRPIN340 suppressed propagation of HIV, herpes simplex virus (HSV) type 1 and 2, Sindbis virus, SARS virus, and cytomegalovirus. These observations have led us to apply SRPIN340 to an antiviral drug.
Furthermore, we are investigating the replication mechanisms of HSV, including how HSV interferes in host gene expression, by analyzing the function of HSV-coding RNA binding proteins.
We have also shown that SRp75 can modulate 5’ splice site selection of Adenovirus E1A gene, and that this activity was enhanced by phosphorylation of SRp75 mediated by Clks. We developed a Clks-specific inhibitor, TG003, and demonstrated that TG003 specifically reduced the phosphorylation level of SRp75. Currently we have been investigating the mechanism of alternative splicing regulation during virus infection and development, in which SRp75 is involved, by analyzing both specific target sequences of SRp75 and the effect of phosphorylation by Clks.
4. Regulation of malignancy specific alternative regulation
Malignant neoplasms account for 13% of total death in the world (WHO, 2006) and thus development of innovative therapies is an urgent and highly prioritized task. In malignancies, multitude of aberrant alternative splicing appear and aggravate their malignant potentials. We hypothesized that novel cancer drug could be invented by targeting the aberrant splicing patterns to more physiological patterns. Our project aim at elucidation of the mechanisms that regulate alternative splicing and targeting them to develop innovative cancer drugs. We are now studying the alternative splicing mechanism of a gene that controls the proliferation of malignant tumors by in cell and in vitro assays. So far, an in vivo splicing imaging reporter was developed that visualizes the switch between different splicing isoforms of the gene in culture cells. We will establish a chemical library screening system using this reporter and seek for novel cancer drugs.
5. Molecular links of mRNA splicing with Transcription coupled DNA repair and mRNA Localization in the cytoplasm
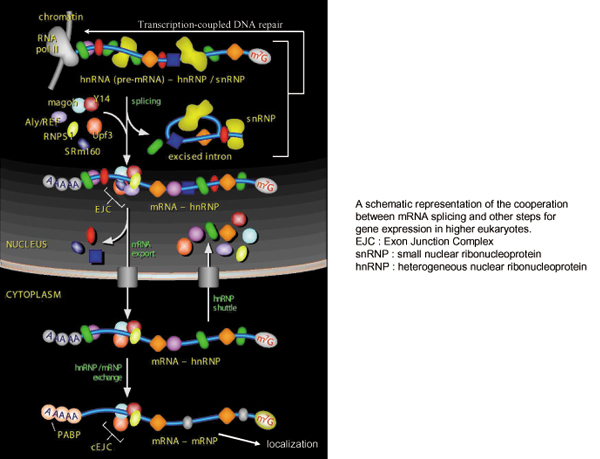
In higher eukaryotes, most genes in the nucleus are separated by introns. Furthermore, more than 95% region of the primary transcripts produced by RNA polymerase II is intronic sequence in human. These results indicate that mRNA splicing is an essential step for gene expression in eukaryotes, and also imply that post splicing turnover of excised introns in the nucleus is important for higher eukaryotes. We would like to understand this step comprehensively including the factors involved. Recently it has been shown that mRNA splicing is a step not only to remove introns but also to make a link with other gene expression steps by depositing Exon Junction Complex near exon-exon junctions. The results we obtained so far suggest that mRNA splicing affects transcription coupled DNA repair and mRNA localization in the cytoplasm of neuronal cells. We would like to uncover new links between mRNA splicing and those steps.
6. Development of Novel Specific Inhibitors of "PSYCHIK" Family Kinases and their Potentials as Pharmaceutical Drugs
Protein kinases are key regulators of wide variety of cellular functions. Inventions of specific protein kinase inhibitors have allowed scientists to elucidate the specific molecular roles of each protein kinases. More importantly, several specific protein kinase inhibitors have been developed as powerful pharmaceutical drugs. The Human Genome Project revealed the total variety of protein kinases; as many as 518 are identified in human genome. For most of them, however, their physiological/pathological roles are poorly studied, and the numbers of clinically useful protein kinase inhibitors are still limited. Therefore, discovery of inhibitors specific for hitherto untargeted kinases are highly anticipated.
We have previously reported the development of SRPIN340, a specific inhibitor of SR protein kinase (SRPK) family, and, TG003, a specific inhibitor of cdc2-like kinase (Clk) family. To date, they are the only specific inhibitors for SRPK and Clk, respectively. Significantly, we also proved that SRPIN340 is a potent anti-viral agent.
We are developing novel inhibitors of other protein kinase families that are phylogenetically related to SRPK and Clk. Being closely related protein kinase families, SRPK and Clk constitute a larger family of phylogenetically related protein kinases: the dual-specificity tyrosine-phosphorylation regulated kinase (DYRK) family, the homeodomain interacting protein kinase (HIPK) family, and pre-messenger RNA processing 4 protein kinase (PRP4). We propose to call the entire family as PSYCHIK family (PRP4, SRPK, DYRK, Clk, HIPK Family). These kinases are considered to engage in important physiological process including the development and normal functioning of the central nervous system, the regulation of apoptosis, and pre-mRNA splicing.
We focus on PSYCHIK family members as targets for discovery of specific inhibitors, and indeed some novel specific inhibitors have been obtained.
We have characterized the new compounds through 1) in vitro assays, 2) in cell functional assays, 3) X-ray crystallography, and 4) whole embryo development assay. The findings demonstrates that the compounds are not only useful biological tools, but are potential drug seeds for hitherto untreatable diseases.
7. Regulation of Dauer Formation (Metamorphosis) in C. elegans.
CREB (cAMP-response element binding protein) is ubiquitously expressed and is involved in multiple biological phenomena such as cell survival, long-term memory and circadian rhythms. CREB is activated by multiple extracellular stimuli via several signaling pathways including cAMP-PKA pathway, MAPK cascade and Calcium/calmodulin (CaM)-dependent protein kinase (CaM-K) cascade. However, signaling pathways leading to CREB activation as well as target genes functioning in the whole animal context are still to be elucidated.
We are currently analyzing phenotypes of the CREB (crh-1) and CaM-K I/IV (cmk-1) mutants. We found that these mutants formed a dauer-like larva, a specialized diapause status of the worm, at high temperature, indicating the involvement of these genes in the dauer regulation pathways. We found that these genes functions in a pair of endocrine cells that produces steroid hormone(s). We are currently searching for signaling cascade leading to CaMK-CREB activation in these cells as well as target genes regulating dauer formation.
|



