

 |
 |
| Contact Us |
Research Interests:
Recent whole genome sequence analyses revealed that a high degree of proteomic complexity is achieved with a limited number of genes. This surprising finding underscores the importance of alternative splicing, through which a single gene can generate structurally and functionally distinct protein isoforms. Based on genome wide analysis, 75% of human genes are thought to encode at least two alternatively spliced isoforms. The regulation of splice site usage, so called gsplicing codeh provides a versatile mechanism for controlling gene expression and for the generation of proteome diversity. Thus splicing code may play essential roles in many biological processes, such as embryonic development, cell growth, and apoptosis. 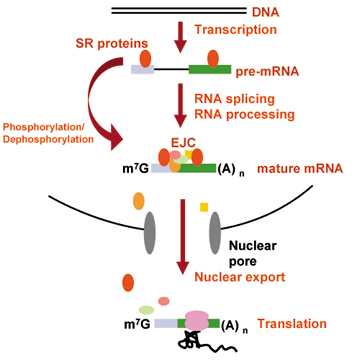 1. Transgenic Reporter Worm System to Monitor Alternative Splicing Patterns in vivo. More than 75% of human genes have alternative mRNA isoforms. What is a spatiotemporal expression profile of each isoform? How are so many genes regulated in vivo? Regulation mechanisms of alternative splicing, however, have been studied mostly in vitro or in cultured cells. We have developed a transgenic alternative splicing reporter system that visualizes expression profiles of 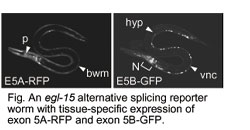 mutually exclusive alternative exons of a nematode C. elegans at a single cell level in vivo (nature methods, 2006). With this reporter system, we have visualized spatiotemporal profiles of tissue-specific and developmentally regulated alternative splicing events in living worms. By isolating and analyzing mutant worms defective in the color profiles, we have 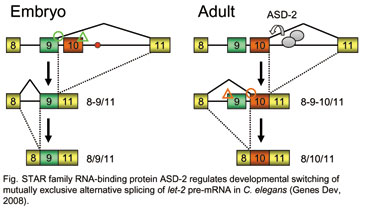 identified trans-acting factors and cis-elements involved in the splicing regulation (Mol Cell Biol, 2007; Genes Dev, 2008). Through these studies, we are coming to realize that molecular mechanisms of the alternative splicing regulation are conserved throughout metazoan evolution. Our reporter system will further elucidate expression profiles and regulation mechanisms of alternative splicing in vivo. identified trans-acting factors and cis-elements involved in the splicing regulation (Mol Cell Biol, 2007; Genes Dev, 2008). Through these studies, we are coming to realize that molecular mechanisms of the alternative splicing regulation are conserved throughout metazoan evolution. Our reporter system will further elucidate expression profiles and regulation mechanisms of alternative splicing in vivo. |
| Copyright (C) 2005-2009 Department of Functional Genomics, Tokyo Medical and Dental University Medical Research Institute & School of Biomedical Science. All Rights Reserved. |