Dr. Liu from China stays in TMDU and NCCRI, Japan
A3 Foresight Program: Education Program for Young Researchers
Report of visit to Department of Molecular Oncology at TMDU and Carcinogenesis Division in NCCRI (Feb. 24-Mar. 2, 2010)
Zhaojun Liu (Department of Cancer Etiology, Peking University School of Oncology)
Host researcher
Prof. Yasuhito Yuasa, M.D Ph.D (Department of Molecular Oncology, Graduate School of Medical and Dental Sciences, Tokyo Medical and Dental University)
Summary
As the education program for young researchers of A3 Foresight Program, I had one week’s visiting stay at Department of Molecular Oncology in Tokyo Medical and Dental University, and Carcinogenesis Division, National Cancer Center Research Institute (NCCRI). The main purpose of this visit was to validate molecular subtyping potential of SRF methylation using Japanese gastric carcinoma samples. I also learned a new bisulfite modification method for DNA methylation analysis during the stay. Besides, I deeply felt the friendship and hospitality of the Japanese people. Bright smile was always seen on their faces. This is a wonderful experience for me and may benefit my future research career.
Contents
1. A novel bisulfite modification of genomic DNA
Bisulfite modification is an important process for methylation analysis. There are number of modification protocols. It takes only two hours by using the MethylampTM One-step DNA Modification Kit in Prof. Yuasa’s lab in Japan, which is different from that used in Prof. Deng’s lab in China. Thus, I learned the time saving method described below.
The procedures are as follows:
Preparation of fresh G1/G2/G3 solution
☆ Add 1.1 ml of G2 solution in the kit into 1 vial of G1 solution before DNA modification. Vortex until the mixture of solutions is clear or saturated. Add 40 µl of G3 solution to the mixed solution, lightly vortex.
Conversion of unmethylated cytosines in DNA
☆ Add 110 µl of the mixed G1/G2/G3 solutions to a 1.5 ml vial containing 10 µl of DNA sample (10 ng to 1 µg). Vortex and place the vial in a thermal cycler with a program of 99C for 6 min followed by 65C for 90 min. The unmethylated cytosines are converted to uridines, but the methylated ones are remained as cytosines after the treatment.
Purification of the modified DNA
☆ Place a spin column into a 2 ml collection tube. Add 300 µl of G4 solution to the column, and then transfer above treated sample to the column. Centrifuge at 12,000 rpm for 20 sec. Remove the column from the collection tube and discard the flowthrough. Replace the column to the collection tube.
☆ Add 200 µl of G5 solution to the column, and centrifuge at 12,000 rpm for 20sec.
☆ Add 10 µl of the G3 solution and 1.1 ml of 90% ethanol into a 1.5 ml vial, mix. Add 50 µl of the mixed G3/ethanol to the column. Sit for 8 min at room temperature, then centrifuge at 12,000 rpm for 20sec.
☆ Add 200 µl of 90% ethanol to the column, centrifuge at 12,000 rpm for 15 sec. Remove the column from the collection tube and discard the flowthrough. Replace the column to the collection tube. Add 200 µl of 90% ethanol to the column, centrifuge at 12,000 rpm for 40 sec.
☆ Put the column in a new 1.5 ml vial. Add 8-18 µl of G6 solution directly to the column filter, centrifuge at 12,000 rpm for 20 sec to elute modified DNA.
2. RT-PCR
We chose 16 gastric or colonic cancer cell lines from Prof. Yuasa’s lab. Total RNA was reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen). After reverse transcription, samples were subjected to amplification of GAPDH.

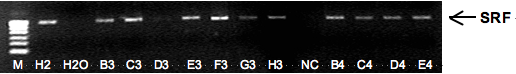
3. Confirmation study on SRF methylation and metastasis of gastric carcinomas
According to our recent work among Chinese patients, methylation of SRF was negatively correlated with metastasis of gastric cancer. It would be more valuable if this result could be confirmed among patients from different countries. So we carried out the validation study using gastric cancer samples from 82 Japanese patients at Prof. Ushijima’s lab at Carcinogenesis Division in NCCRI. The SRF CpG island was amplified with the same CpG–free primer set I used in China. The DNA samples of gastric carcinomas were extracted and modified with bisulfite ready before my visit. Since a single band of SRP PCR product was obtained from 78 samples, methylation status of SRF in these samples was analyzed by DHPLC. A significantly negative correlation between SRF methylation and metastasis of gastric carcinomas from Japanese patients was observed. Taken together, those results indicate that SRF methylation may be a potential biomarker for prognosis of gastric carcinomas of patients. A joint manuscript will be written soon.
4. Presentation of my previous work
I gave a presentation of my work ‘Detecting DNA methylation biomarkers for gastric cancer metastasis’ in the Department of Molecular Oncology on Mar. 1. Prof. Yuasa, Dr. Akiyama, and other laboratory members gave me some good suggestions about my work.
Acknowledgments and a photo
I thank Dr. Akiyama for showing me their bisulfite-modification technique and other experimental skills, Ms Ozawa for giving me a great help in my daily life, Dr. Hattori & Ms. Koba for taking care of me in the NCCRI, Dr. Otsubo & Ms. Mimata & Ms. Rotkrua & Mr. Hashimoto for introducing the Japanese culture to me, and all laboratory members for their warm hospitality, respectively. I had a wonderful time with all of you.
I sincerely thank Prof. Yuasa for kind acceptance and all arrangement about my visit to TMDU. I also thank Prof. Ushijima for our productive collaborative work.




