HOME > Networking > Dr. Wang from China stays in NCCRI, Japan
Dr. Wang from China stays in NCCRI, Japan
A3 Foresight Program: Education Program for Young Researchers
Report of visit to National Cancer Center Research Institute in Japan (Feb.24-Mar.3, 2010)
Xi Wang (Department of Biochemistry and Molecular Biology, Peking University Health Science Center)
Host researcher
Prof. Toshikazu Ushijima, Ph.D. (Carcinogenesis Division, National Cancer Center Research Institute)
Summary
By attending the education program for young researchers of A3 foresight program, I studied in Prof. Ushijima’s laboratory from Feb.24 to Mar.3, 2010. During this period, I got a general view of advanced technologies in DNA-methylation analysis and the role of specific gene methylation in gastric cancer carcinogenesis. I also focused on the workflow of quantitative real-time MSP assays. I deeply appreciate this chance to visit Prof. Ushijima’s laboratory. I got great experiences and wonderful memories from Japanese colleagues and friends.
Contents
Experiments
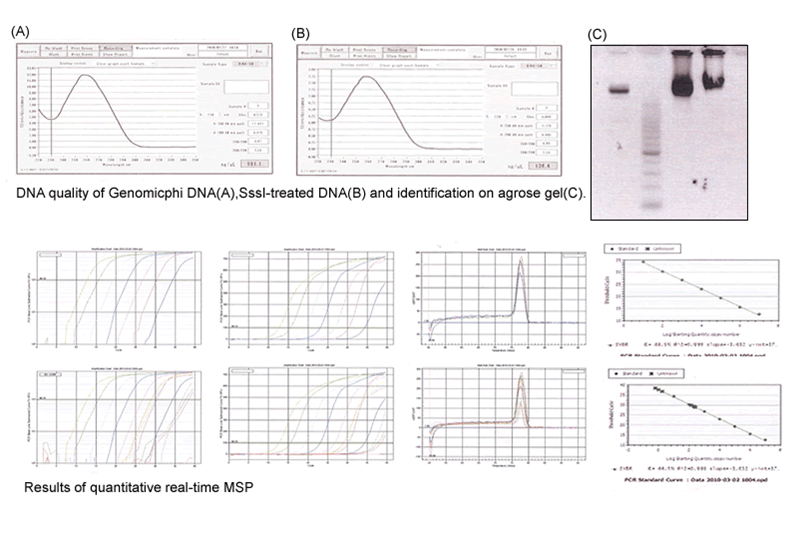
1. Overview of quantitative real-time MSP assays
Real-time MSP is a quantitative analysis of methylated or unmethylated DNA at a specific region. Numbers of methylated or unmethylated DNA molecules can be quantified by comparing their amplification with the amplification of standard DNA. The methylation level can be calculated based on the number of DNA molecules.
2. Preparation of control samples
• Preparation of unmethylated DNA from GenomiPhi Kit
• Denaturation of template DNA
• DNA amplification via reaction system containing input DNA, dNTPs, random hexamer primers and Phi29 DNA polymerase
• BamHI digestion and purification
3. Preparation of methylated DNA by SssI treatment
• 1st DNA modification by SssI via reaction system containing BamHI digested DNA, SssI enzyme, SAM and reaction buffer
• DNA purification by phenol/chloroform
• 2nd DNA modification in the same condition as 1st treatment
• DNA purification by phenol/chloroform
4. Sodium bisulfite treatment
• DNA denaturation
• Sodium bisulfite treatment via cycling reaction system containing NaHSO3, NaOH and hydroquinone
• DNA purification by Zymo-Spin column
5. Quantitative real-time methylation specific PCR with standard DNA and sample DNA by gene specific primers
Communications
1. Introducing and discussing my current work and research interests in Journal Club on Feb.25
2. Discussing experiments design and technical details with Dr. Hattori
3. Communicating and interchanging ideas about projects with Dr. Takeshima, Dr. Asada, Dr. Yoshida and Dr. Ando
Acknowledgement
I sincerely thank Prof. Ushijima for kindly and warm reception. I thank Dr. Hattori not only for the thoughtful arrangement and patient instruction of experimental technologies but also for the warm concern in my daily life. I thank Ms. Kobayashi, Ms. Mori, Ms. Wakabayashi and all other lab members for the generous help and warm hospitality. It’s my great pleasure to meet you all in Tokyo.



