Dr. Otsubo from Japan stays in KRIBB, Korea
A3 Foresight Program: Education program for young researchers
Report of visit to KRIBB in Korea (Oct. 25 - Nov. 1, 2009)
Takeshi Otsubo (Department of Molecular Oncology, Graduate School of Medical and Dental Sciences, Tokyo Medical and Dental University)
Host researcher
Prof. Yong Sung Kim, Ph.D (Medical Genomics Research Center, KRIBB)
Summary
As the education program for young researchers of A3 Foresight Program, I visited to Prof. Yong Sung Kim's laboratory (Medical Genomics Research Center, KRIBB) in Korea. The main purpose of this visit was to learn about a high-throughput methylome assay system "MeDIP-seq" technique and many ideas about the research on epigenomics in gastric cancer. Actually, during my stay in KRIBB, I learned about not only the techniques and new knowledge but also the Korean culture and the importance of interpersonal communication. My visit to KRIBB has been a great experience for the rest of my researcher life, because I have got a lot of insight into new technologies and foreign friends.
Contents
1.Overview of the MeDIP-seq technique
Methy-DNA immunoprecipitation (MeDIP) is an immunocapturing approach based on the direct immunoprecipitation of methylated DNA fragments. In the MeDIP-seq analysis, the methylated DNA fragments are then directly sequenced by using a high-throughput sequencing technology. This technique is the excellent method for identifying methylated CpG-rich sequencing at a genome-wide level.
2.Preparation of the MeDIP samples
This is the most important step of MeDIP-seq analyses, because if the samples are not properly prepared, we cannot obtain good results reproducibly.
The procedures are as follows:
> Nebulize (Fragment) the purified DNA
> MeDIP with MBD-Biotin Protein (invitrogen, MethylMinerTM Methylated DNA Enrichment Kit)
> Preparation for Solexa sequencing (illumina, ChIP-Seq Sample Prep Kit)
- Perform End Repair
- Add 'A' Bases to the 3 End of the DNA Fragments
- Ligate Adapters to DNA Fragments
- Size Select the Library
- Enrich the Adapter-Modified DNA Fragments by PCR
- Validate the Library
Representative Results of the Preparation of the MeDIP samples
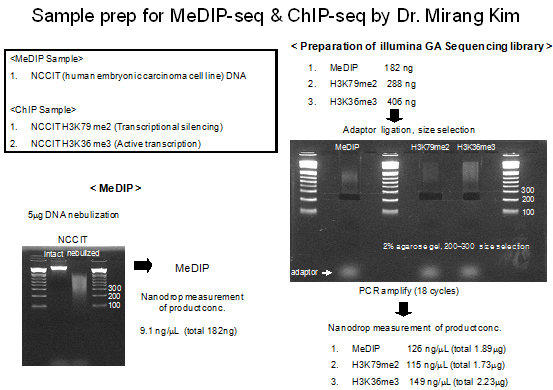
3.Overview of the high-throughput sequencer "Solexa Genome Analyzer II"
Prepared MeDIP samples can be directly sequenced by Solexa Genome Analyzer II (illumina). The Genome Analysis System process is consisting of following four steps:
- Cluster generation on the Cluster Station
- Sequencing-by-Synthesis (SBS) on Genome Analyzer
- Paired-End Module
- Data analysis using the Genome Analyzer Pipeline software (CASAVA)
4.Presentation of my own work
I introduced my own work "Epigenetic silencing of SOX2 by DNA methylation and microRNA in gastric cancers" and my interests in this program at KIRBB on Oct. 27.
Photos and acknowledgment

I thank Dr. Mirang Kim for instructing the experimental techniques of MeDIP-seq, Dr. Han-Chul Lee for picking up me to KRIBB on the first day, Mr. Kwun Ill Kim & Ms. Kee Ok Ham for helping my stay in Korea, and all laboratory members for the warm hospitality, respectively. I sincerely thank Prof. Yong Sung Kim for kindly acceptance and all arrangement about my visit to KRIBB.



